Targeting the 3D epigenome in pediatric sarcomas.
I am a doctoral student in the lab of Dr. Berkley Gryder. One main goal of our research is to investigate epigenetic mechanisms controlling 3D folding of the genome and transcriptional activity. Targeted cancer treatments often depend on dysregulated signaling axes, but in many other cancer types that urgently need new treatments, the whole transcriptional landscape is shifted to enable high levels of the transcriptional machinery that promote expression of many different oncogenes. In addition, genomic rearrangements can also affect chromatin states and create new enhancer interactions that can enable gains-of-function such as colonization of a new tissue type.
My area of interest within the lab is to apply insights into epigenetic regulation towards fusion-positive rhabdomyosarcoma (RMS), a pediatric soft tissue sarcoma with a transcription factor fusion that prevents terminal differentiation by creating novel super-enhancers around oncogenes, and osteosarcoma (OS), a bone cancer that tends to undergo complex genomic rearrangements and metastasize to the lung. In samples from both diseases, we have performed mapping of 3D chromatin contacts using HiChIP and histone marks such as H3K27ac, and we are using RNA-seq data to correlate these findings with gene expression. In RMS, we have included samples treated with drugs targeting acetylation machinery, while in OS, we have included primary and metastatic pairs to investigate lung colonization. I aim to build upon prior work and preliminary data to understand disease-state-specific 3D contacts that can be unraveled into key dependencies, with broad therapeutic implications for cancer types with similarly dysregulated epigenomic and transcriptional states.
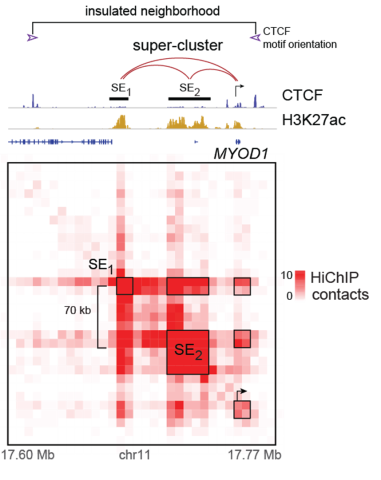
Figure (Left): 2D ChIP-seq tracks of CTCF (defining topologically associated domains) and H3K27ac (defining open chromatin) above a 3D contact matrix of H3K27ac-mediated connections at MYOD1 super-enhancers in a fusion-positive RMS cell line.
Figure (above): Approaches for collaborative investigation of OS metastasis through in vivo metastasis models, 3D chromatin profiling, rational chemical design, and in silico modeling of cell competencies over time.