I am currently a doctoral trainee in Dr. Yan Li's lab. We are interested in applying functional genomic, epigenomic, and single cell genomic tools to study islet biology, beta cell development, and the genetics of complex diseases such as obesity and diabetes. Type II Diabetes (T2D) is a metabolic disorder characterized by dysfunction of insulin-secreting beta cells in the pancreatic islet. Loss of insulin results in an inability to regulate blood glucose levels. The risk factors for T2D are well-reported, however there is still much we do not know about the mechanism behind how these risk factors contribute to T2D disease development. As T2D rates continue to rise in the United States, understanding the intricacies of pancreatic islet gene expression can help us better answer these questions.
My work is focused on characterizing how T2D risk factors such as age, sex, and blood glucose levels affect beta cell gene expression and chromatin accessibility profiles. We’ve gathered a cohort of dozens of human pancreatic islets from both healthy and diabetic donors. Using the single cell RNA-seq platform, Drop-seq, and a combinatorial indexing method of single nuclei ATAC-seq, we have identified gene expression and chromatin accessibility changes associated with age, diabetic status, HbA1C levels, as well as sex-differences in beta cell expression. We’ve further expanded our analyses to look at other cell types in the pancreas to better understand their role in the diabetic environment including alpha, delta, and pancreatic stellate cells. It is our hope that this dataset could shed some light on diabetes pathogenesis and serve as a useful resource for islet biologists.
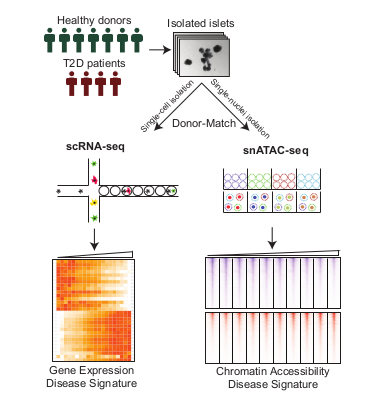
Project Workflow
Healthy and diabetic islets from human donors were collected and processed for donor-matched single cell RNA-seq using microfluidics platform, Dropseq, and single nuclei ATAC-seq using a combinatorial indexing platform. Differentially expressed genes and accessible chromatin regions were then identified, characterizing the genomic and epigenomic profile of disease-relevant genes in pancreatic islets.