I am a Systems Biology and Bioinformatics doctoral trainee with Dr. William Bush, in the department of Population and Quantitative Health Sciences. Our lab focuses on understanding the functional impact of genetic variation, with specific emphasis on studying genetic drivers of Alzheimer’s disease.
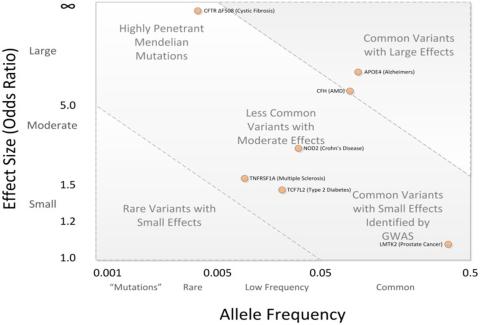
Alzheimer’s disease (AD) is genetically complex and known to be associated with a wide range of genetic factors. Thus far over 70 genetic loci associated with disease risk, disease outcomes, and risk for specific AD subtypes have been identified. However, the genetic factors discovered so far account for only a portion of the underlying genetic architecture. Advancements in sequencing technologies and expansions of study cohorts have generated data with much higher genomic fidelity, allowing us to study classes of variation that have not been well-explored.
My projects are focused on rare variants and structural variants. Rare genetic variants typically have larger effect sizes on disease risk compared to common variants, as well as bigger impacts on protein function. However, current gene-based tests for rare variants generally consider the impact of low-frequency coding variants as an independent effect from the more commonly occurring regulatory variants that surround them. We show that we can increase the statistical power of kernel-based rare-variant association tests by incorporating common variants around a gene of interest as a fixed effect. This method is then applied to a large genetic dataset of over 34,000 individuals from the Alzheimer’s Disease Sequencing Project to find new gene candidates for analyses.
Structural variants are another class of variants that have not been systemically explored, despite representing the largest and most complex source of human genomic diversity. We intend to use the long-read data to characterize the structural variation of haptoglobin, which has complex subtypes unable to be resolved by short-read sequencing. Using haptoglobin as a prototype, further analyses of long-read data aim to identify novel and rare structural variants which current reference genomes and variant calling tools are not trained to detect.
Rare variants and structural variants are major sources of unobserved genomic diversity that have yet to be well-characterized and tested for involvement in Alzheimer’s disease. This proposal aims to integrate multiple sequencing technologies to understand the genetic basis of complex traits and discover new associations to treatment and disease.